Excessively reproduced research: Detection of plagiarism in PubMed abstracts
1. Introduction
This post reports on a small project for finding evidence of plagiarism in PubMed abstracts.
The scientific method is a somewhat curious endeavor. You want explicitness and transparency for data, methods, and results. If you didn't have those, it would be very hard to do science because in almost all cases we build on prior work. You might apply a new method to existing data or you might get some new data and apply an existing method to it. You might even try to reproduce somebody's work exactly using exact data and exact methods. But when all is said and done, you're supposed to make an entirely original presentation of your work. When you don't, that's plagiarism.
Here's a definition from Wikipedia. I am citing it explicitly because only a huge idiot would plagiarize the definition of plagiarism.
Plagiarism is the representation of another author's language, thoughts, ideas, or expressions as one's own original work.1
2. Prior work
Here's a brief review of plagiarism in medical scientific research papers. Their abstract starts with a definition of plagiarism. You might be having a hard time reading that abstract because of the large red word printed across it.

This article was retracted because it was stolen from: Mohammed, R.A., Shaaban, O.M., Mahran, D.G., Attellawy, H.N., Makhlof, A. and Albasri, A., 2015. Plagiarism in medical scientific research. Journal of Taibah University Medical Sciences, 10(1), pp.6-11.
There really is a fine line between stupid and clever.2
2.1. Crossref
Most of the plagiarism detection systems employ some kind of fuzzy matching algorithm. For example, Crossref, from Elsevier, uses a system called ``iThenticate'' to identify duplicates. It's proprietary, so hard to get meaningful numbers.

2.2. My goal
My goal in this project was very simple: find all the sentences in PubMed abstracts that appear more than once? I required an exact, character-for-character match. I wanted to do this for many abstracts (millions).
3. Methods
3.1. Data
The data are the approximately last 8 years of PubMed XML files (up to 2018). The way PubMed works is that they release new batches in XML format. But they're not just the latest publications. They also include updates to prior publications, and each entry in the XML file contains a version number. Therefore some of the entries in a recent file could be updates of earlier publications. It's a little bit difficult to say exactly what the date range I used, but roughly speaking, it's the last eight years.
Here's a cartoon version of my algorithm. I make a dictionary (hash table) that points from each sentence to a list of abstracts containing that sentence. To carve up each abstract into sentences, I use a rather complex regular expression. It doesn't work perfectly, but that doesn't matter because it's deterministic. The same abstract going through each time will get parsed the same way.
Basically I'm making an inverted index from sentences to all the abstracts that contain those sentences.
3.2. Step 1
index = dict()
for each abstract
for each sentence
index[sentence] += abstract
For the second step, find all sentences that occur in more than one abstract.
3.3. Step 2
for each (sentence, abstracts) in index
if length(abstracts) > 1
record sentence as possible suspect
The actual code is much longer and more complex. To get this to run on my laptop, because the XML files are rather large, the code makes multiple passes over the data to reduce the storage space needed. (This also speeds the processing.) All of these details have been omitted here. If you had enough RAM and storage for a database, you could implement something very close to my cartoon algorithm.
3.4. Heuristic clean-up
Although requiring an exact character-for-character match sets a very high threshold of detection, there are still false positives. I introduced a set of heuristics to remove some kinds of false positives:
- Remove sentence if occurs in more than 3 abstracts
- Remove sentence if too short
- Remove abstract if it is a ``Reports an error in''or ``Reports the retraction of''
- Remove abstract if it has fewer than 3 sentences
- Remove sentence if abstract appears in journal that prints abstracts from other journals as part of a commentary
- Remove pairs of abstracts sharing a sentence if there are any authors in common
I will try to motivate at least some of these heuristics. If a sentence occurs in a lot of abstracts, it's not stolen, it's boilerplate text, probably something as simple as a copyright notice. If you follow the PubMed rules, copyright notices aren't supposed to appear in the abstract, but some publishers don't follow these rules.
Really short sentences, just by virtue of their length, have a relatively high probability of recurring if you process many abstracts; these sentences are just not that interesting.
Really short abstracts are also not interesting; there's just not that much information within.
Some abstracts appear in article entries that are pointers to other entries in these XML files. So the abstract is an partial or exact duplicate. One way this happens is when entries are linked by ``reported error in'' or ``report the retraction of''.
Another issue is self-plagiarism. There are people who sometimes publish the same thing more than once. In certain circumstances, that's completely legitimate. In other cases, it's highly frowned upon, and I didn't want to get into trying to adjudicate this issue. So I decided to adopt a liberal policy that if there's a non-null intersection between the author lists of two suspect abstracts, then that's acceptable. I'm only retaining the abstract pair when there's complete lack of overlap in the authors.
4. Results
The size of the dataset is shown in this table:
| XML Files | 200 |
|---|---|
| Abstracts | 5,070,585 |
| Sentences | 41,365,959 |
The processing produces 14,460 pairs of abstracts, such that each member of the pair shares at least one sentence with the other member of the pair.
Because I don't have any ground truth labels of plagiarism, I don't know how many true positives there are, or how many false positives there are. Since what I really want to do is find ethical violations, it's going to require manual post-processing stage to confirm because you're not going to have an automated system posting candidate plagiarism without a very careful human review.
4.1. What are the false positives?
As noted earlier, sometimes things like copyright information appear widely, and sometimes the second article is a commentary, and shares the same abstract.
My shared-author heuristic doesn't work in some circumstances, such as for the Spanish naming custom of having two last names. Depending upon which last name is listed as the last name of the author my heuristic might fail.
In some countries, sometimes one's last name is written first; the journal may or may not represent that correctly. If they don't do the right thing then my heuristic fails.
There are certainly entirely acceptable circumstances in which, for example, clinical guidelines are published simultaneously in multiple journals.
And it seems like at least within the community of people who do meta analyses, it may be legitimate to start with a previous meta analysis, particularly, if it was conducted by your team and then updated five or 10 years later. If so, then some of that text might be identical.
4.2. What are the true positives?
What are the real true positives? It looks very likely that they are text used without attribution; in some cases this applies to entire articles.
Here are some examples. These were handpicked by me to illustrate some points. Automatically picking high-likelihood candidates is not a problem I have solved.
What I produce is a large HTML file; this is so that it can be easily viewed in a browser, which allows reformatting of size and width, and also supports hypertext links.
4.3. Example 1
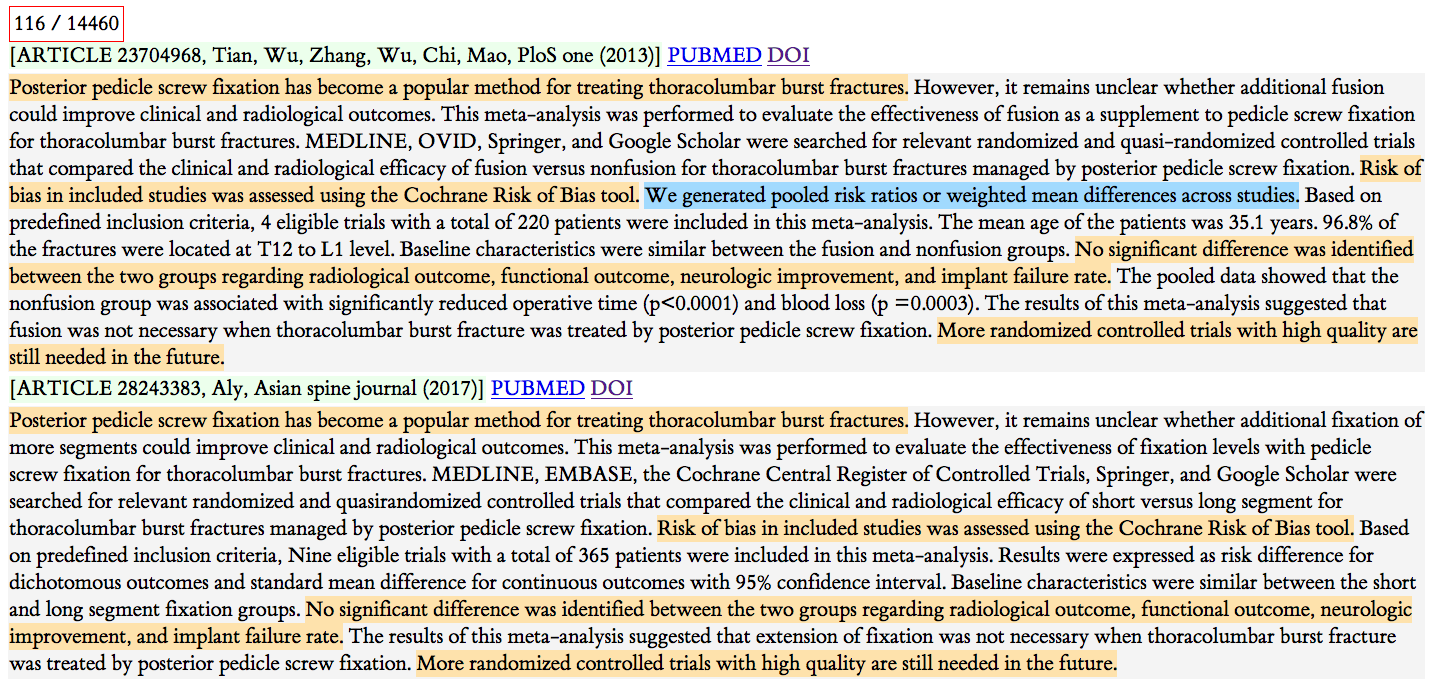
Figure 1: Example 1
What we see here is a pair of articles. Each article's entry starts with the light green background. That line shows the PubMed ID, the list of authors, the journal name, a link to the PubMed entry, and the DOI link for the actual article. The second article in the pair is below and appears in the same format.
The colors indicate sharing. This yellow sentence starting ``Posterior pedicle screw fixation ….'' is exactly the same sentence in both abstracts. You can see there are some other short shared sentences as well.
The light blue marks a sentence that is shared with a different abstract that is not a member of this pair.
My hypothesis for what happened here is that the second article (from 2017) used without attribution text from the first article (from 2013). The second article does not cite the first one. Both are meta analyses, but with slightly different foci. I suspect the second author just borrowed some text because he was lazy. Or he maybe was in a rush.
4.4. Example 2
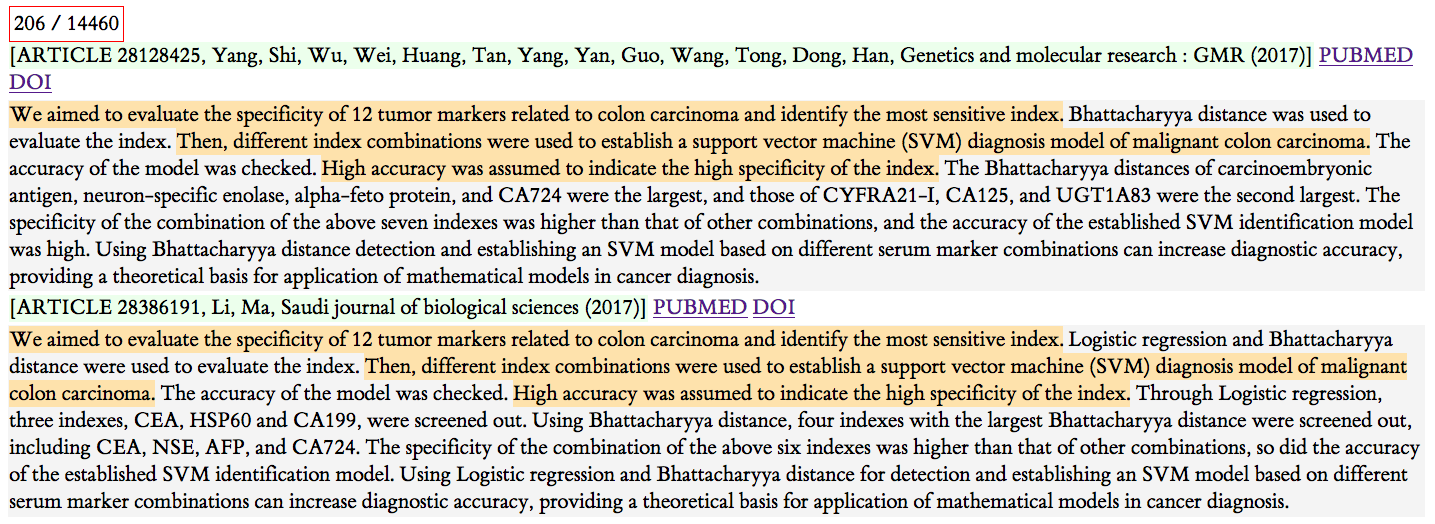
Figure 2: Example 2
Here we have another pair of articles, with different authors, different journals, and the same year. A couple sentences are exactly the same, but it's not entirely clear when you read the rest of the abstract what's going on. If you dive into the article text, you find that it is nearly entirely identical.
Note that it is not clear in this instance who borrowed from whom. Check out these articles and decide for yourself: https://pubmed.ncbi.nlm.nih.gov/28128425/ and https://pubmed.ncbi.nlm.nih.gov/28386191/
They did change the name of the hospitals where the research was conducted. They also have different conclusions. My hypothesis is that the ``borrower'' maybe ran up against a word limit and had to shorten their conclusion. I don't really know. Note that they have different acknowledgments and list different funding sources.

Figure 3: Yang et al 2017

Figure 4: Li et al 2017
4.5. Example 3
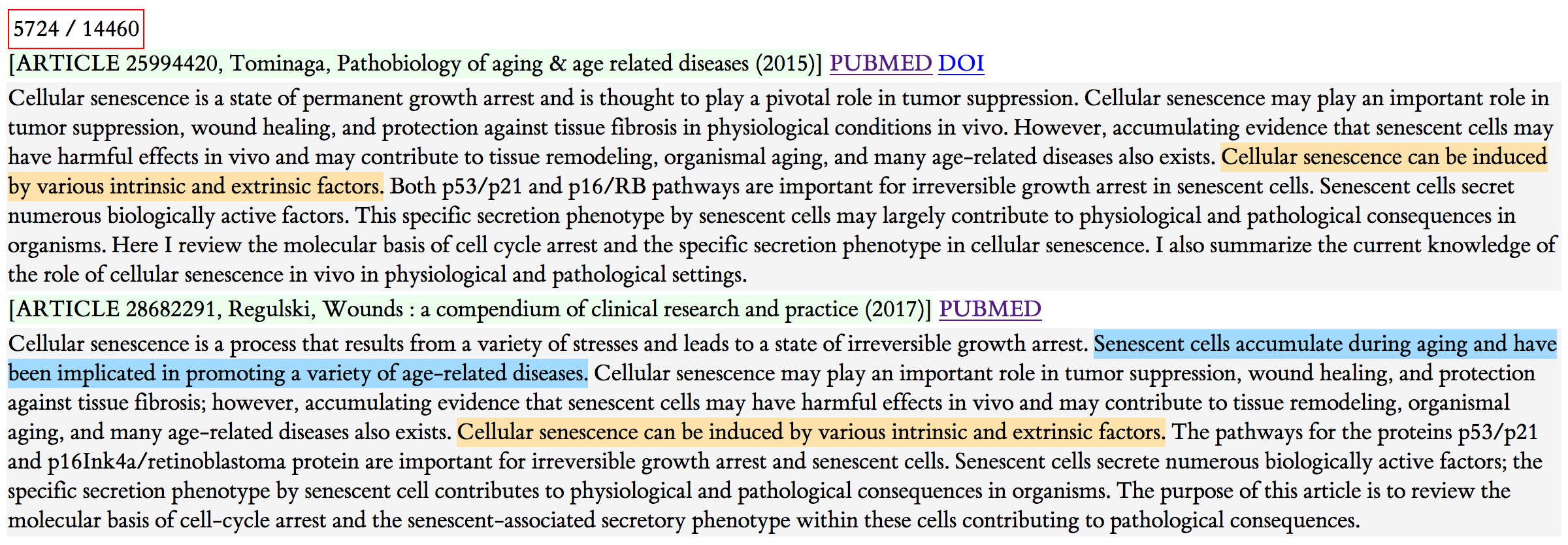
Figure 5: Example 3
In this third example abstract, there's only one sentence that's shared, although some of the couple of sentences look similar.
This one is very suspicious. The articles were published two years apart. In the article text, there are many borrowed sentences. The figure below shows one such example which I've marked by hand; the sentences vary by one word, and the references 20–23 are the exact same references as 39–42 in the second paper. Note that Regulski's paper does not cite Tominaga's earlier paper. With some investigation, I found out that Regulski is on the advisory board of the journal that published this paper.

Figure 6: Tominaga 2015

Figure 7: Regulski 2017
5. Why do people do this?
I think authors are under time pressure or the pressure to increase one's publication count. The authors have an urgent need for some background text, or, in the more flagrant examples, entire manuscripts.
Perhaps they are not fluent in English. It's easy to adapt previously published text.
I don't think there is much scrutiny, or much of an enforcement mechanism. My hypothesis is that some journal editors aren't strongly motivated to police manuscripts—you're just going to get into a dust-up with some authors and it's just not worth the hassle. If you go after particularly influential scientists or people at prestigious universities, they have a lot of power and it's not clear who's going to win that game. You hope that truth would win but truth sometimes takes a beating.
6. What to do?

The above is from THE, which is the Times of London Higher Education supplement, basically pointing out that some systemic things like regulatory institutional reforms could help and highlighting the value of having important researchers remain close to and aware of what's happening in their part of science, as opposed to moving up to positions where they're essentially just managers.
How about explicitly teaching ethical behavior? That sounds like a good idea. It turns out it doesn't work very well. Here's an interesting Stanford panel from 2014, "Does teaching ethics do any good". The professors who are participating in the panel, who were in fact educators of ethics, agreed that ethics classes cannot be expected to make students more ethical.

… the Stanford professors who participated in the discussion agreed that ethics classes cannot be expected to make students more ethical.
Eric Schwitzgebel, a philosopher at UC Riverside, has done some work in this area and basically says the same thing in a couple different studies that ``we should be aware that this assumption is dubious and without good empirical support.'' – the assumption being that there's a positive influence of this teaching on the moral behavior.
… if part of the justification is an assumed positive influence on students’ moral behavior after graduation, we should be aware that that assumption is dubious and without good empirical support.''2
6.1. What does influence ethical behavior?
There's various personality factors, codes and mores, the dynamics of groups, and how they respond to such things. Laws and regulations, and culture also matter, and incentives certainly have effects on behavior, ethical and otherwise.

Figure 8: Nature, Vol 579, 5 March 2020
Just recently, China banned cash rewards for publishing. Obviously China is a very large country, and there are many scientists in China, who may have been given improper incentives to publish and perhaps publish things that maybe weren't even theirs. That China has now at least removed one of these incentives is a good sign.
6.2. What is the role of informatics?
I did not use a probability model to determine how likely a sentence is. This depends a lot on the sentence length and the content of sentence. But I did not try to unleash the awesome power on machine learning. I was doing something very simple.
If you had reasonable output from a classification system, followed by a stage for human assessment, you could start doing some interesting things, looking for patterns by author, by institutions, by country, by journals, by journal types, or by impact factors. But without accurate classification this is hard to do, but we had that we could say some interesting things about the current state and maybe help shape policy.
The fact that somebody is on the editorial board and is also feels perfectly comfortable adapting text from other articles which he doesn't even bother to cite, and publishing it in a journal he has a strong affiliation with—it might be nice to see where all these people lie within a social network.
6.3. Possible projects
If you have access to a server and you have all the PubMed abstracts on that server, you could do something as straightforward as that very cartoon version of the algorithm.
Ideally, this would be part of a larger workflow, where the new XML updates would come in, you'd register them to database, reconcile different versions of these things, run some code and then have a team of people who reviewed positives by hand and then the fun begins. You then email the journal editors of the implicated articles and say, ``Hey, I look at which I found. What do you think?''. If they don't get back to you with three or six months, you post everything on a website of miscreants.
I think NLM should also consider doing something like with all submissions to PubMed. Or Google—they already have an inverted index of most of the Web.
Finally, there's a very interesting blog, called Retraction Watch, that I highly recommend, although it's a bit discouraging. They track retractions of scientific publications. Some of them are real ethical miscues—they are just people who have stolen content or made stuff up. In other cases, it is a bit harder to tell. And if what I propose became a real thing, I think that Retraction Watch would be very interested in hearing about the results.
7. Acknowledgments
This blog post is a revision of a talk I gave April 7, 2020 to Stanford Biomedical Informatics, as part of series of ethics discussions for its trainees. Thanks to Jaap Suermondt of BMI for the opportunity to present and for the audience's great comments and questions.
Footnotes:
Source: Plagiarism - Wikipedia
Source: This is Spinal Tap